Pu•MA System 3D for Spheroid Toxicity Testing and In Situ Imaging
Introduction
There is an increasing interest in using three-dimensional (3D) cell structures for modeling tumors, organs, and tissue to accelerate translation research1 . Significant progress has been made in formation of such structures to recapitulate the in vivo environment but performing complex assays with them can be challenging. For example, manual treatment, staining, and processing of spheroids and organoids is typically labor-intensive and prone to disruption or loss of samples. Here we report on use of our microfluidic-based Pu·MA® System to perform automated toxicity assays with spheroids. Spheroids can be incubated with and without compounds for 1 to 5 days and then analyzed in situ by high content imaging. Other cell viability methods such as measurement of ATP are also feasible.
Pu·MA System Workflow
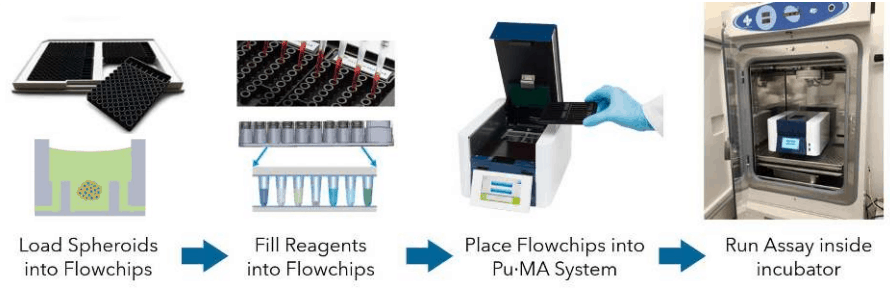
Pu·MA System Flowchips are designed with chambers and reservoirs arranged in a convenient multi-well plate format (384-well spacings) and provide up to 32 tests per plate. Once spheroids and reagents are loaded into the flowchips, the plate is placed into the Pu·MA System and reagent exchanges are done automatically through microfluidic channels connected to the protected sample chamber (Fig 1). Multiple reagent exchanges can be performed enabling complex assay protocols to be run.
Assay programs are pre-loaded into the system and run using an intuitive touch-screen interface. The whole Pu·MA System can be placed in an incubator to run assays at 37°C and 5% CO2. The system architecture and use of pneumatics to move fluids provides gas exchange to the sample chambers.
Figure 1. Schematic of the Pu·MA System workflow.
Spheroid Toxicity & Imaging
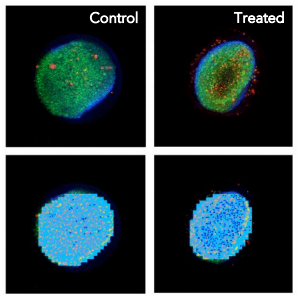
Capabilities of the system were demonstrated by assaying cellular toxicity over a 24-hour period with HCT116 spheroids. The spheroids were incubated in media with and without compound. The flowchips have a clear imaging window at the bottom of the sample chamber to enable high resolution imaging of the cell structures.
Assay Procedure
- Colon cancer cells (HCT116) cells (~2K cells) were grown and incubated for 48 hours until they formed tight spheroids.2
- The spheroids were transferred to a Pu·MA System flowchip in either media (Control) or media + compound (Treated) and placed into a Pu·MA System in an incubator.
- After compound incubation, spheroids were stained and washed prior to imaging.
- Imaging was done with an ImageXpress Micro Confocal Imaging System and analysis performed using the Custom Module Editor with MetaXpress Automated Imaging Acquisition and Analysis Software.
Cell Viability Assay
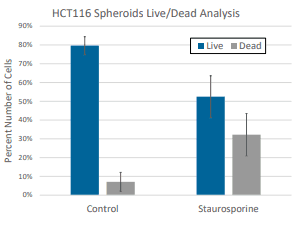
Cell viability was measured for HCT116 spheroids incubated with compounds for 48 hours in Pu·MA System flowchips. (Compounds: C59 30 mM; Mitomycin 30 mM; Etoposide 100 mM). Amount of ATP present in each spheroid sample was measured using the CellTiter-Glo assay (Promega). 10 µl of CellTiter-Glo reagent was automatically transferred into the sample wells and incubated for 10 min. Luminescence signal was measured using a SpectraMax iD5 Plate Reader (Molecular Devices) (Fig 4).
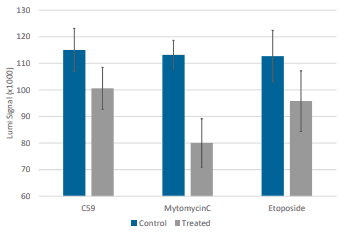
Conclusions
- We have demonstrated capabilities of a novel automated 3D Cell-based assay system that performs complex protocols with spheroids in an incubator environment.
- The Pu·MA System has been shown to perform in situ lysing of spheroids to provide highly sensitive metabolomic profiling with minimal perturbation.
- The ability to analyze spheroids and organoids in situ in order to capture toxicity information and metabolomic profiles shows great promise for oncology research.
References
- Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning, Langhans, S.A. Frontiers in Pharma. 2018, 9, 1.
- High-Content Assays for Characterizing the Viability and Morphology of 3D Cancer Spheroid Cultures, Sirenko, O. et al. Assay and Drug Dev Tech. 2015, 13, 402.
Pu·MA System Microfluidics
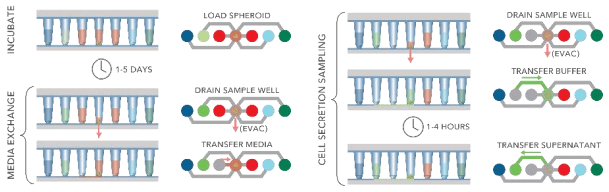
The Pu·MA System can automatically perform complex assay steps using proprietary microfluidics. All reagents are loaded into flowchips and then incubation, media exchanges, cell secretion sampling and other steps are executed by the system program (Fig 5 and 6).
- 3D Cell models remain in the protected in bottom chamber
- Can exchange up to 95% of media without drying cells
- No direct fluid flow over cells to disrupt cell structures
Figure 6. Schematic of fluid transfer in flowchip sample well. A spheroid in well is shown on left.
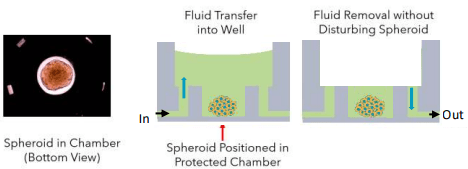
Figure 5. Schematic showing typical protocol steps available with the Pu·MA System. All steps are performed automatically inside an incubator.
Repeatability
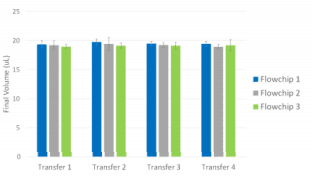
Fluid transfer repeatability was demonstrated by exchange of multiple buffers back and forth between Pu·MA System flowchip wells (Fig 7).
- 20 µl fluid transfers in 8 lanes, 3 repeats
- Fluid transferred to Well 3, incubated for 30 min, and then transferred back to origin well.
- Volume in origin well was measured gravimetrically
- Final Volume CV = 3%
Figure 7. Fluid transfer demonstration results. Error bars represent +/- 1 SD (n=8)