Automated Biomarker Identification using IF Staining using the Pu·MA System® followed by High-Content Confocal Imaging
Introduction
Physiologically relevant 3D cell models are essential for pre-clinical research and drug discovery because they better represent physiological processes and disease phenotypes 1,2 . Biomarker studies have become a cornerstone in preventive and personalized medicine. There is an increased interest in using cellular biomarkers to complement genomic ones 3,4 . Both development of functional assays and identification of cellular biomarkers in 3D models are critical, as they are expected to have a higher clinical importance than in 2D culture systems 5. Immunofluorescence staining (IF) is a widely used method to identify and quantify biomarker expressions and their cellular localizations. One of the challenges faced by researchers is that IF staining of delicate spheroids/organoids tend to be manual and tedious.
Culture systems like hanging drop or ultra-low attachment are difficult to handle for IF staining due to a risk of losing or damaging the spheroids. There is a further technical challenge of plate compatibility and locating spheroids for confocal microscopy, which is typically used for imaging of stained spheroids. The commonly used approach of embedding, and sectioning is cumbersome, time consuming and prone to errors. To overcome these challenges, we have developed and describe here a breakthrough advancement in biomarker staining and detection. We present the automated IF staining in 3D cell models using our proprietary microfluidic-based Pu·MA System 4,6,7 combined with high resolution confocal imaging technology from Yokogawa Electric Corporation (Figure 1).
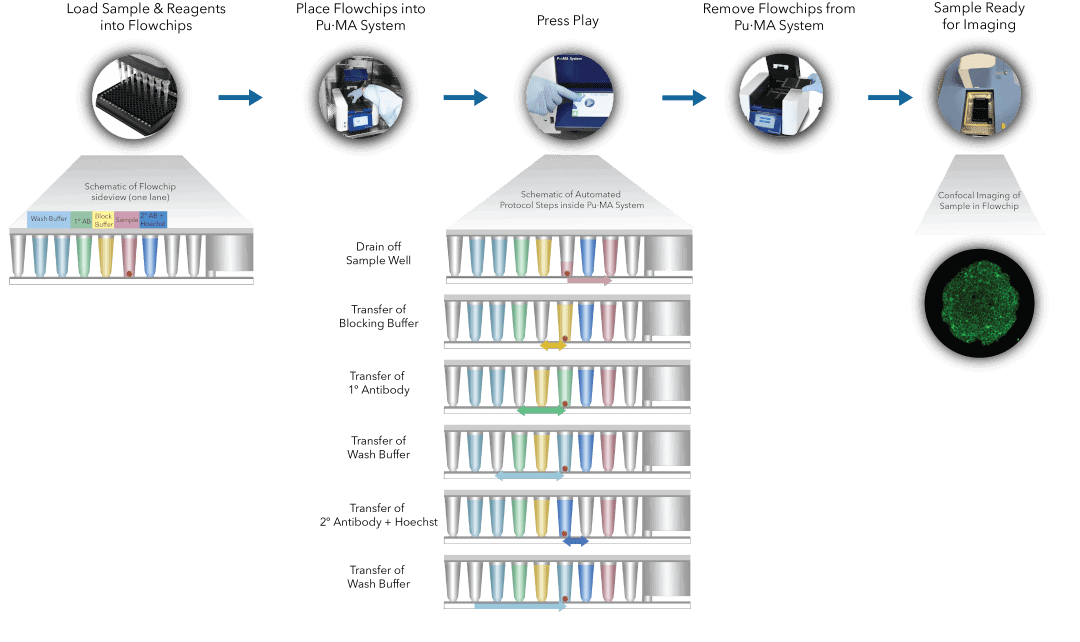
Fig 1. Pu·MA System IF staining workflow schematic representation of automated steps within the Pu·MA System followed by subsequent confocal imaging within flowchips
