Automated Dynamic In Situ Sampling for Metabolite Analysis of Breast Cancer Tumoroids
Introduction
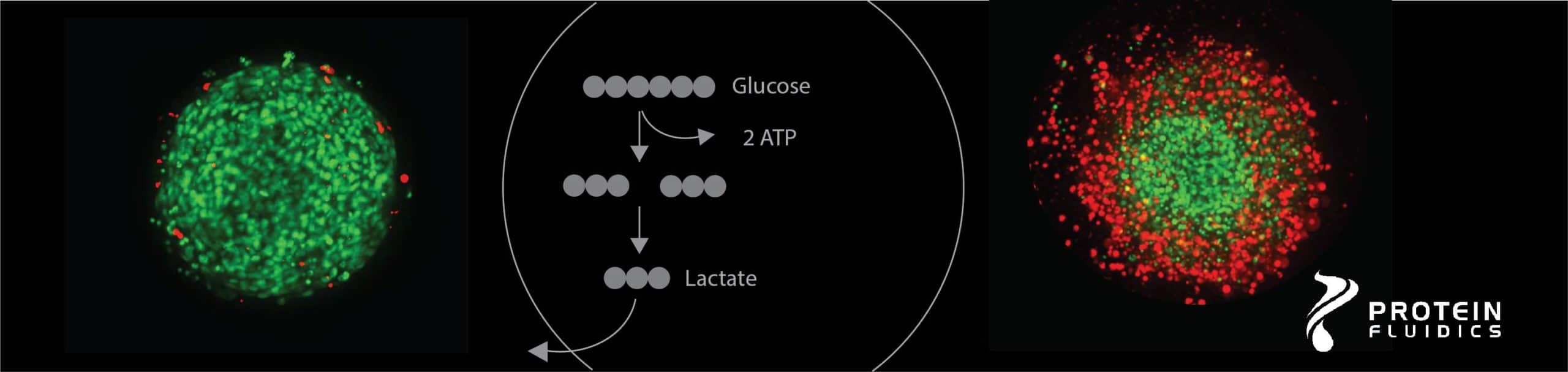
Cancer cells undergo metabolic modifications to meet high bioenergetic and anabolic demand during tumorigenesis. For most tumors, aerobic glycolysis is a major driver of cancer progression, resistance to chemotherapy, and poor patient outcome. Lactate, a product of glycolysis, plays a role in tumor progression and is linked to drug resistance in breast cancer. Patient-derived 3D cancer cell models are valuable tools for research, drug development and personalized medicine, because they recapitulate features of the tumor microenvironment. Therefore, monitoring metabolite dynamics and lactate production in physiologically relevant patient-derived models can provide valuable data for understanding metabolic perturbations during disease progression, drug response and resistance.
In this technote, we demonstrate a workflow to assess metabolic changes in triple negative breast cancer (TNBC) tumoroids in response to treatment with anti-cancer drugs. Our approach utilizes the Pu·MA System with microfluidic flowchips that allows for dynamic in situ supernatant sampling, followed by a sensitive luminescence lactate assay and high content imaging.
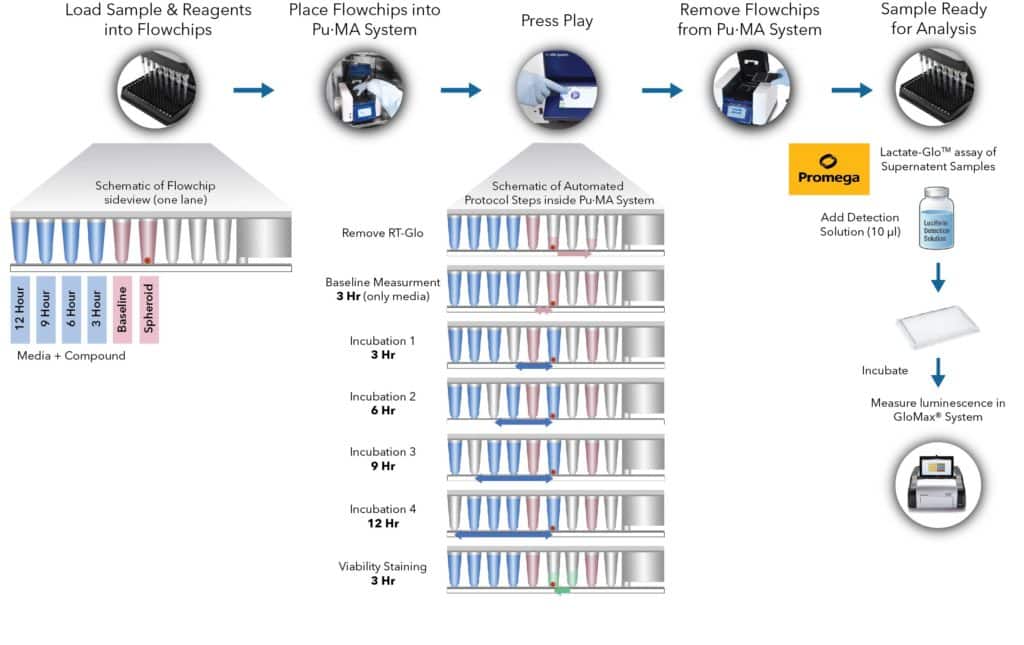
Fig 1. Metabolite secretion assay workflow with schematic representation of automated steps within the Pu·MA System. This is followed by supernatant analysis for the secreted metabolite lactate.
